PCR Reactions
Due to the individual conditions required for optimal PCR of fluorescently labeled fragments (especially if multiplexing is being used), we are unable to perform your PCR reactions for you. We are also unable to tell you what dilution of your products will "work" for you. There is no simple correlation between a dilution ratio and how your fragments will appear on the run. Our experience does, however, tell us that it is almost always better to perform some form of dilution of the PCR reactions prior to mixing with the size marker and running on the analyser. This may only be 10-20x or it may be 200-400x. The range is quite large.
To assist with optimisation of dilutions, we are more than happy to run a test plate for you. We will take a selection (e.g. 8) of your samples and run these at serial two fold dilution, usually starting at 1:10. This means you will get back 48 results with dilutions ranging from 1:10 to 1:320. We can provide an "overview" of how these results look, but, for obvious reasons, we are not able to tell you exactly which dilution is best for you. The test plates are charged at the same rate as normal plates even though it is considerably more work for us. Please contact us should you wish to discuss this further.
Analysis of Results
We will undertake limited quality control analysis of all runs to include checking that the marker bands are present and well defined and that sample injection has occurred in all capillaries. This is included in the basic cost of the work.
We are afraid that we cannot offer to undertake a full analysis of your results and produce allele information.
Please note that customers can make use of free software available from Applied Biosystems called PeakScanner for their analysis and display of the data.
96-well Plate Recommendations
You are free to send us your samples in any standard 96 well plate of your choice. However, please note that it must be a standard 96 well plate containing 12 columns of 8 wells. Please ensure it is clear which corner is the "A1" well and note that we always work in columns of 8 and NOT rows of 12! When sending 48 or less samples, also please note that our 48 capillary machine is set up to take samples from the odd columns (1,3,5,7,9,11) and so your samples should be put into those columns.
When sealing the top of the plate, there are various methods you can use such as heat sealing, adhesive film or a silicon sealing mat. We recommend only using a silicon mat as in our experience adhesive film does not stick well and can result in leakage out of wells. Also, heat sealing is difficult for us to remove in order to access the samples. For customers wishing to use silicon mats, we recommend "Axymats" from Axygen. The one we use has product code "AM-96-PCR-RD". We will return mats in batches to customers for re-use. Whichever sealing method you choose, please ensure it is effective. We don't like having to tell customers that their sealing did not work and all the wells have been mixed/emptied of contents....
For those customers who only require a small number of plates and do not wish to buy a whole box, please contact us and we will supply you with the number of plates you need at £10 per plate (including postage & packing) along with a protective base plate and a silicon sealing mat to keep your samples in the wells when you return the plate together with your samples.
Organization of Samples on Plate
- You may send either 48 or 96 samples per plate.
- Please note that even if you send less than 48 or 96 samples, we will need to charge you for the full 48 or 96 samples.
- Please organize the samples into the wells in the format given below.
- Failure to do this will result in the samples not being processed.
- Please ensure that you know where each of your samples is on the plate, since result files will be numbered as illustrated below (i.e. consecutively from 1...48 or 1...96).
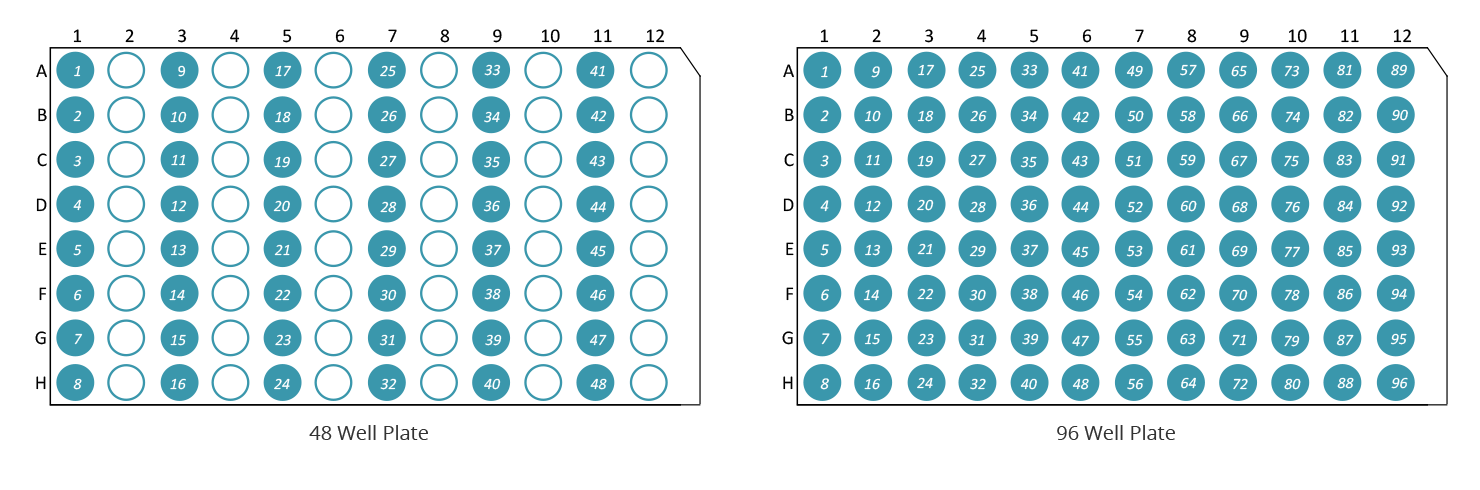
- Please note the cut corner on the plate (upper right) and the reference numbers/letters that will also appear on the microtitre plate.
- Please ensure that your samples are placed exactly as per the illustrations above or you will not receive your samples back in the correct order.
Once all your samples are ready, seal the plate and send to us.
 Fragment Analysis, a type of genetic marker analysis, is a technique that facilitates identifying the presence or absence of a DNA sequence that is linked to an allele of interest. It does not provide the sequence information of a particular gene; instead changes in the length of specific DNA sequences provide key information to distinguish changes in markers. Often, analyses such as these are used to examine inheritance patterns of alleles of interest in families (for instance, those associated with a disease) and even populations – including humans, animals and plants.
Fragment Analysis, a type of genetic marker analysis, is a technique that facilitates identifying the presence or absence of a DNA sequence that is linked to an allele of interest. It does not provide the sequence information of a particular gene; instead changes in the length of specific DNA sequences provide key information to distinguish changes in markers. Often, analyses such as these are used to examine inheritance patterns of alleles of interest in families (for instance, those associated with a disease) and even populations – including humans, animals and plants.

